Network Medicine Institute and Global Alliance
Representing 33 leading universities and institutions around the world committed to improving global health and advancing the field of Network Medicine

What is network medicine?
Network Medicine combines principles and approaches from network sciences, systems biology, and human dynamics to understand the causes of human diseases and develop new treatments.
History of Network Medicine
Network Medicine evolved from network science research during the early days of the internet and advances in systems biology since the human genome project. Now it's an established way to study, reclassify, and develop treatments for complex diseases.
1991
CERN introduces the World Wide Web to the public.
1999
Barabasi introduced the concept of scale-free networks and proposed the Barabási–Albert model to explain their widespread emergence in natural, technological and social systems. Barabasi’s paper on Scale-Free Networks in Science Magazine becomes the most-cited paper in the Physical Sciences.
2003
The Human Genome Project was declared complete in April 2003. The project determined there are approximately 22,300 protein-coding genes in human beings. Loscalzo and Barabasi begin building the map of human disease biology that explains how proteins expressed from the human genome interact to cause specific diseases.
2007
Loscalzo, Kohane, and Barabási publish Human disease classification in the postgenomic era in the Molecular Systems Biology journal which establishes Network Medicine as a complex systems approach to human pathobiology.
2011
Barabási, Gulbahce, and Loscalzo publish Network Medicine: A Network-based Approach to Human Disease in Nature where they present an overview of the organizing principles that govern cellular networks and the implications of these principles for understanding disease.
2012
The Channing Division of Network Medicine at Brigham and Women's Hospital was created to study, reclassify, and develop treatments for complex diseases using network science and systems biology.
2016
Joseph Loscalzo and Enrico Petrillo form the Network Medicine Alliance representing 31 leading universities and institutions around the world.
2017
Joseph Loscalzo, Albert-László Barabási, and Edwin Silverman publish the seminal Network Medicine textbook.
2018
Joseph Loscalzo and Albert-László Barabási publish Network-based approach to prediction and population-based validation of in silico drug repurposing which demonstrated that a unique integration of protein-protein interaction network proximity and large-scale patient-level longitudinal data complemented by mechanistic in vitro studies can facilitate drug repurposing.
The Transformation of Medicine, The First International Conference on Network Medicine and Big Data was held in Rome, Italy. The ultimate goal of the meeting was to design a strategy by which this interdisciplinary field can truly transform medicine.
2019
The Channing Division of Network Medicine (CDNM) continues to expand its staff of more than 80 Harvard Medical School faculty and 42 fellows in addition to 160 non-faculty Brigham and Women’s Hospital (BWH) employees. BWH oversees the second largest hospital-based research program in the world. CDNM’s annual research expenditures represented 25% of the BWH Department of Medicine annual budget. In fiscal year 2019, CDNM investigators received 54 new funding awards resulting in 173 active grants.
2020
In response to the COVID-19 pandemic, the Network Medicine Institute launches the COVID-19 Global Drug Repurposing Study (GDR Study). The GDR Study is based on a multifaceted drug repurposing strategy which includes a global patient registry and an advanced Network Medicine framework to identify promising therapies among the thousands of already FDA approved drugs. The strategy also facilitates evidence-based updates to health policy and clinical guidelines and tailors them to specific patient populations.
Selected Major Network Medicine Alliance consortium Grants 2022:

REPO4EU was funded with a highly competitive 5-year Twenty-Five Million Euro grant from the European Union Governments Horizons Research Program.
It is developing a comprehensive European and Global platform for precision drug repurposing applying existing drugs to be repurposed for diseases with significant unmet needs via innovative treatment protocols. The core ofREPO4EU consists of a team of world-leading scientists with breakthroughs in advanced bioinformatics and artificial intelligence (AI) on real-world big data to redefine diseases in a mechanism-based manner. This revolutionary new era of medicine will allow unprecedented efficacy and cost- effectiveness in treating diseases globally. Within 5 years, REPO4EU will establish a first-in-class coherent and innovative web-based platform for safe and efficient drug repurposing for all types of high unmet medical need indications to all European researchers and SMEs with a unique Open Science concept, ensuring global medical impact.
2021-2022 Research Highlights
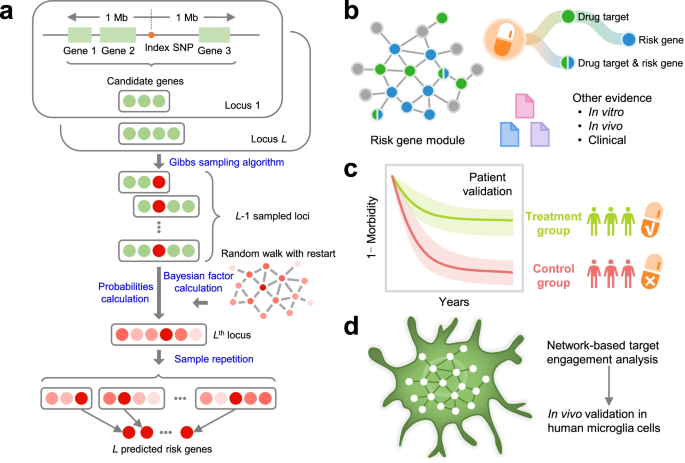
Genome-wide association studies (GWAS) have identified numerous susceptibility loci for Alzheimer’s disease (AD). However, utilizing GWAS and multi-omics data to identify high-confidence AD risk genes (ARGs) and druggable targets that can guide development of new therapeutics for patients suffering from AD has heretofore not been successful.
We developed an endophenotype disease module-based methodology for Alzheimer’s disease (AD) drug repurposing and identified sildenafil as a potential disease risk modifier. Based on retrospective case–control pharmacoepidemiologic analyses of insurance claims data for 7.23 million individuals, we found that sildenafil usage was significantly associated with a 69% reduced risk of AD (hazard ratio 0.31, 95% confidence interval 0.25–0.39, P < 1.0 × 10–8). Propensity score-stratified analyses confirmed that sildenafil is significantly associated with a decreased risk of AD across all four drug cohorts tested (diltiazem, glimepiride, losartan and metformin) after adjusting for age, sex, race and disease comorbidities. We also found that sildenafil increases neurite growth and decreases phospho-tau expression in neuron models derived from induced pluripotent stem cells from patients with AD, supporting mechanistically its potential beneficial effect in AD. The association between sildenafil use and decreased incidence of AD does not establish causality, which will require a randomized controlled trial.
Traditional drug discovery faces a severe efficacy crisis. Repurposing of registered drugs provides an alternative with lower costs and faster drug development timelines. However, the data necessary for the identification of disease modules, i.e. pathways and sub-networks describing the mechanisms of complex diseases which contain potential drug targets, are scattered across independent databases. Moreover, existing studies are limited to predictions for specific diseases or non-translational algorithmic approaches. There is an unmet need for adaptable tools allowing biomedical researchers to employ network-based drug repurposing approaches for their individual use cases. We close this gap with NeDRex, an integrative and interactive platform for network-based drug repurposing and disease module discovery. NeDRex integrates ten different data sources covering genes, drugs, drug targets, disease annotations, and their relationships. NeDRex allows for constructing heterogeneous biological networks, mining them for disease modules, prioritizing drugs targeting disease mechanisms, and statistical validation. We demonstrate the utility of NeDRex in five specific use-cases.
Cardiovascular disease is a leading cause of death in general population and the second leading cause of mortality and morbidity in cancer survivors after recurrent malignancy in the United States. The growing awareness of cancer therapy–related cardiac dysfunction (CTRCD) has led to an emerging field of cardio-oncology; yet, there is limited knowledge on how to predict which patients will experience adverse cardiac outcomes. We aimed to perform unbiased cardiac risk stratification for cancer patients using our large-scale, institutional electronic medical records.
Dementia-like cognitive impairment is an increasingly reported complication of SARS-CoV-2 infection. However, the underlying mechanisms responsible for this complication remain unclear. A better understanding of causative processes by which COVID-19 may lead to cognitive impairment is essential for developing preventive and therapeutic interventions.
The COVID-19 pandemic has highlighted the need to quickly and reliably prioritize clinically approved compounds for their potential effectiveness for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections. Here, we deployed algorithms relying on artificial intelligence, network diffusion, and network proximity, tasking each of them to rank 6,340 drugs for their expected efficacy against SARS-CoV-2. To test the predictions, we used as ground truth 918 drugs experimentally screened in VeroE6 cells, as well as the list of drugs in clinical trials that capture the medical community’s assessment of drugs with potential COVID-19 efficacy. We find that no single predictive algorithm offers consistently reliable outcomes across all datasets and metrics. This outcome prompted us to develop a multimodal technology that fuses the predictions of all algorithms, finding that a consensus among the different predictive methods consistently exceeds the performance of the best individual pipelines. We screened in human cells the top-ranked drugs, obtaining a 62% success rate, in contrast to the 0.8% hit rate of nonguided screenings. Of the six drugs that reduced viral infection, four could be directly repurposed to treat COVID-19, proposing novel treatments for COVID-19. We also found that 76 of the 77 drugs that successfully reduced viral infection do not bind the proteins targeted by SARS-CoV-2, indicating that these network drugs rely on network-based mechanisms that cannot be identified using docking-based strategies. These advances offer a methodological pathway to identify repurposable drugs for future pathogens and neglected diseases underserved by the costs and extended timeline of de novo drug development.
Polyphenols, natural products present in plant-based foods, play a protective role against several complex diseases through their antioxidant activity and by diverse molecular mechanisms. Here we develop a network medicine framework to uncover mechanisms for the effects of polyphenols on health by considering the molecular interactions between polyphenol protein targets and proteins associated with diseases. We find that the protein targets of polyphenols cluster in specific neighbourhoods of the human interactome, whose network proximity to disease proteins is predictive of the molecule’s known therapeutic effects. The methodology recovers known associations, such as the effect of epigallocatechin-3-O-gallate on type 2 diabetes, and predicts that rosmarinic acid has a direct impact on platelet function, representing a novel mechanism through which it could affect cardiovascular health. We experimentally confirm that rosmarinic acid inhibits platelet aggregation and α-granule secretion through inhibition of protein tyrosine phosphorylation, offering direct support for the predicted molecular mechanism. Our framework represents a starting point for mechanistic interpretation of the health effects underlying food-related compounds, allowing us to integrate into a predictive framework knowledge on food metabolism, bioavailability and drug interaction.
Technological and computational advances in genomics and interactomics have made it possible to identify how disease mutations perturb protein–protein interaction (PPI) networks within human cells. Here, we show that disease-associated germline variants are significantly enriched in sequences encoding PPI interfaces compared to variants identified in healthy participants from the projects 1000 Genomes and ExAC. Somatic missense mutations are also significantly enriched in PPI interfaces compared to noninterfaces in 10,861 tumor exomes. We computationally identified 470 putative oncoPPIs in a pan-cancer analysis and demonstrate that oncoPPIs are highly correlated with patient survival and drug resistance/sensitivity. We experimentally validate the network effects of 13 oncoPPIs using a systematic binary interaction assay, and also demonstrate the functional consequences of two of these on tumor cell growth. In summary, this human interactome network framework provides a powerful tool for prioritization of alleles with PPI-perturbing mutations to inform pathobiological mechanism- and genotype-based therapeutic discovery.
New Members to the Network Medicine Alliance in 2021-2022
Medical University of Vienna (MedUni Vienna) is one of the most traditional medical education and research facilities in Europe. With almost 8,000 students, it is currently the largest medical training center in German-speaking countries. With 6,000 employees, 30 departments and two clinical institutes, 13 medical theory centers, and numerous highly specialized laboratories, it is also one of Europe's leading research establishments in the biomedical sector.
Paris-Saclay University is a public research university based in Paris, France. It is one of the 13 prestigious universities that were produced from the division of the University of Paris, known as the Sorbonne.
The Max Perutz Labs are dedicated to a mechanistic understanding of fundamental biomedical processes. By analyzing and reconstituting complex biological systems across different scales, our scientists aim to link breakthroughs in basic research to advances in human health.
The Complexity Science Hub Vienna is a Vienna-based research organization with the aim to bundle, coordinate and advance the research of complex systems, system analysis and big data science in Austria.
The University of Padua is one of Europe’s oldest and most prestigious seats of learning. As a multi-disciplinary institute of higher education, the University aims to provide its students with professional training and a solid cultural background. The qualification received from the University of Padua act as a symbol of the ambitious objectives respected and coveted by both students and employers alike.
Cyprus Institute of Neurology & Genetics
The Cyprus Institute of Neurology & Genetics (CING) is a private, non-profit, bi-communal, medical, research and academic center.
The INSTITUTE
The Institute's mission is to provide resources and guidance to alliance members so their research can have a global impact on health policy and clinical guidelines and untap the full potential of personalized medicine.
The Institute is a nonprofit with entities recognized by the United States and European Union.

THe Alliance
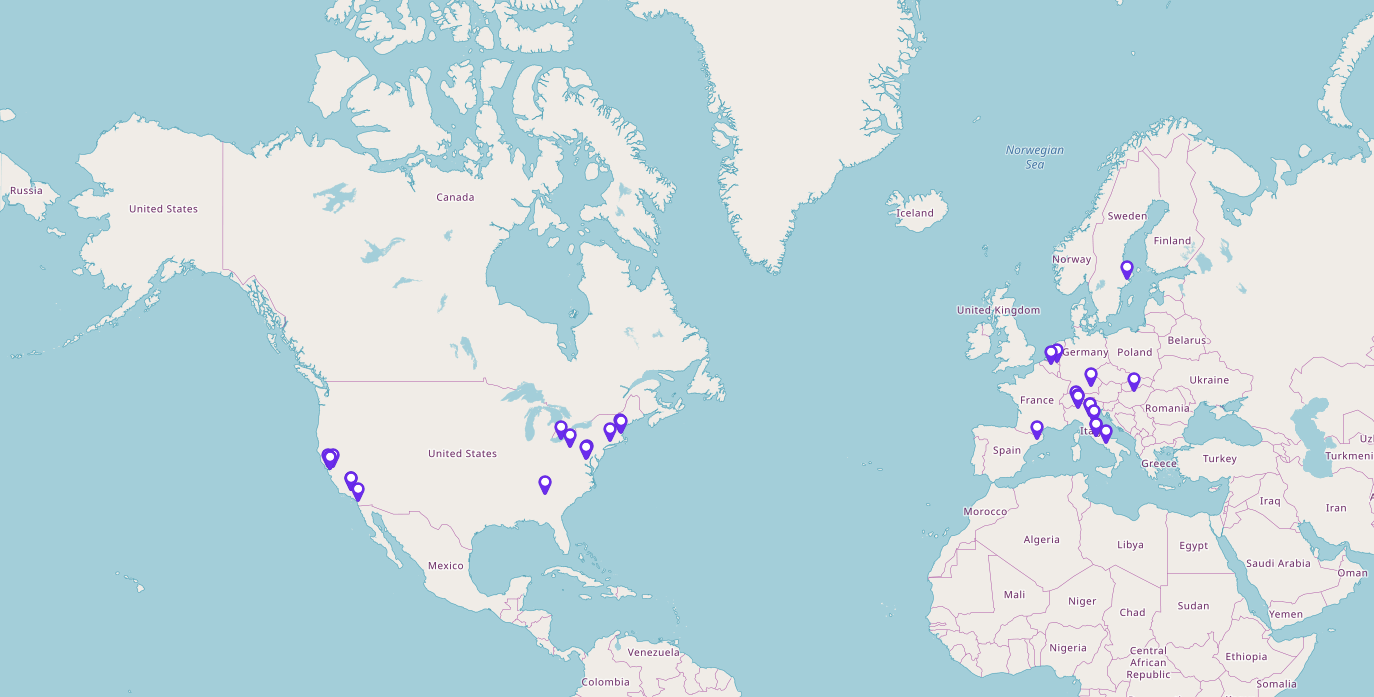
The Network Medicine Alliance represents 33 leading universities and institutions around the world. It enables members to share expertise and best practices while also providing leverage to influence health policy and clinical guidelines.
Selected Publications
We clustered gene expression and methylation data in 78 lung tissue samples from former smokers with normal lung function or severe COPD. We applied two integrative omics clustering methods: (1) Similarity Network Fusion (SNF) and (2) Entropy-Based Consensus Clustering (ECC).
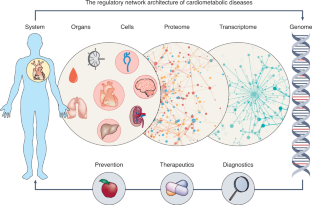
Scientific reductionism has been the basis of disease classification and understanding for more than a century. However, the reductionist approach of characterizing diseases from a limited set of clinical observations and laboratory evaluations has proven insufficient in the face of an exponential growth in data generated from transcriptomics, proteomics, metabolomics and deep phenotyping. A new systematic method is necessary to organize these datasets and build new definitions of what constitutes a disease that incorporates both biological and environmental factors to more precisely describe the ever-growing complexity of phenotypes and their underlying molecular determinants. Network medicine provides such a conceptual framework to bridge these vast quantities of data while providing an individualized understanding of disease. The modern application of network medicine principles is yielding new insights into the pathobiology of chronic kidney diseases and renovascular disorders by expanding the understanding of pathogenic mediators, novel biomarkers and new options for renal therapeutics. These efforts affirm network medicine as a robust paradigm for elucidating new advances in the diagnosis and treatment of kidney disorders.
Despite impressive efforts invested in epigenetic research in the last 50 years, clinical applications are still lacking. Only a few university hospital centers currently use epigenetic biomarkers at the bedside. Moreover, the overall concept of precision medicine is not widely recognized in routine medical practice and the reductionist approach remains predominant in treating patients affected by major diseases such as cancer and cardiovascular diseases. By its’ very nature, epigenetics is integrative of genetic networks. The study of epigenetic biomarkers has led to the identification of numerous drugs with an increasingly significant role in clinical therapy especially of cancer patients. Here, we provide an overview of clinical epigenetics within the context of network analysis. We illustrate achievements to date and discuss how we can move from traditional medicine into the era of network medicine (NM), where pathway-informed molecular diagnostics will allow treatment selection following the paradigm of precision medicine.
Environmental factors, and in particular diet, are known to play a key role in the development of Coronary Heart Disease. Many of these factors were unveiled by detailed nutritional epidemiology studies, focusing on the role of a single nutrient or food at a time. Here, we apply an Environment-Wide Association Study approach to Nurses’ Health Study data to explore comprehensively and agnostically the association of 257 nutrients and 117 foods with coronary heart disease risk (acute myocardial infarction and fatal coronary heart disease). After accounting for multiple testing, we identify 16 food items and 37 nutrients that show statistically significant association – while adjusting for potential confounding and control variables such as physical activity, smoking, calorie intake, and medication use – among which 38 associations were validated in Nurses’ Health Study II. Our implementation of Environment-Wide Association Study successfully reproduces prior knowledge of diet-coronary heart disease associations in the epidemiological literature, and helps us detect new associations that were only marginally studied, opening potential avenues for further extensive experimental validation. We also show that Environment-Wide Association Study allows us to identify a bipartite food-nutrient network, highlighting which foods drive the associations of specific nutrients with coronary heart disease risk.
Network Medicine is a scientific discipline that focuses on the interaction between biological components, such as proteins, microRNAs, or metabolites, to understand molecular pathways that underlie the pathogenesis of diseases. More recently, Network Medicine has expanded to integrate molecular data with phenotypic features as a means by which to clarify mechanisms driving clinical disorders. Since the first publication introducing Network Medicine in 20071, nearly 3300 scientific reports have advanced and refined this discipline, which aims to use scientific “big data” to decipher the role of molecular interactions in the context of health and human disease.
The new discipline of Network Medicine stems from the growing realization that conventional scientific reductionism is inadequate for dissecting complex diseases, increasing efficacy of prevention strategies, or tailoring precise therapies. In addition, Network Medicine acknowledges that health and disease must be viewed in the context of the interplay among multiple molecular and environmental determinants that must be fully considered in precision diagnostics and therapeutics. Network Medicine, therefore, aims to use innovative technology, information, and big data to create an integrated set of principles and discoveries that can fully capture these inherent dependencies, are relevant and translatable to the clinic, and, consequently, are truly innovative in their implementation. The principles and discoveries aim to influence prevention, diagnosis, and treatment beneficially. The focus should be, for example, toward shifting cancer health care management from diagnosis and treatment (extremely expensive) to prevention (the highest cost/benefit ratio); treating patients with chronic noncommunicable diseases (eg, cardiometabolic diseases) more effectively by understanding how the underlying pathogenic processes act, for example, in different age groups; and comprehending the keys for healthy aging across the life course. Drug development may also be enhanced by Network Medicine. Conventional drug development is limited to a relatively small number of targets that are highly expensive to characterize, not clinically precise, and time-consuming to analyze and test. Repositioning of existing drugs can function more economically to minimize costs, time, and risk. Network Medicine offers a useful platform to address all of these challenges.
The Visual Analytics cycle applied to Network Medicine. Data from different domains (e.g., cellular, molecular, and genetic networks) are input to two different processes, Visual Data Exploration which exploits visualization paradigms (Node‐Edge, Matrix, Chords, etc.) to represent these data and classic Automated Data Analysis through different approaches (machine learning, network analysis algorithms, etc.). These two processes are interconnected, allowing an analyst to steer algorithms by interacting with the visual representation of results. The whole process generates new insights (e.g., relationships among networks) used as a feedback loop for new cycles of analysis.
First International Conference on Network Medicine and Big Data
September 24–26, 2018
Rome, Italy
Abstracts
Joseph Loscalzo, MD, PhD
Despite extensive evidence showing that exposure to specific chemicals can lead to disease, current research approaches and regulatory policies fail to address the chemical complexity of our world. To safeguard current and future generations from the increasing number of chemicals polluting our environment, a systematic and agnostic approach is needed. The “exposome” concept strives to capture the diversity and range of exposures to synthetic chemicals, dietary constituents, psychosocial stressors, and physical factors, as well as their corresponding biological responses. Technological advances such as high-resolution mass spectrometry and network science have allowed us to take the first steps toward a comprehensive assessment of the exposome. Given the increased recognition of the dominant role that nongenetic factors play in disease, an effort to characterize the exposome at a scale comparable to that of the human genome is warranted.
Our understanding of how diet affects health is limited to 150 key nutritional components that are tracked and catalogued by the United States Department of Agriculture and other national databases. Although this knowledge has been transformative for health sciences, helping unveil the role of calories, sugar, fat, vitamins and other nutritional factors in the emergence of common diseases, these nutritional components represent only a small fraction of the more than 26,000 distinct, definable biochemicals present in our food—many of which have documented effects on health but remain unquantified in any systematic fashion across different individual foods. Using new advances such as machine learning, a high-resolution library of these biochemicals could enable the systematic study of the full biochemical spectrum of our diets, opening new avenues for understanding the composition of what we eat, and how it affects health and disease.
Despite exceptional experimental efforts to map out the human interactome, the continued data incompleteness limits our ability to understand the molecular roots of human disease. Computational tools offer a promising alternative, helping identify biologically significant, yet unmapped protein-protein interactions (PPIs). While link prediction methods connect proteins on the basis of biological or network-based similarity, interacting proteins are not necessarily similar and similar proteins do not necessarily interact. Here, we offer structural and evolutionary evidence that proteins interact not if they are similar to each other, but if one of them is similar to the other’s partners. This approach, that mathematically relies on network paths of length three (L3), significantly outperforms all existing link prediction methods. Given its high accuracy, we show that L3 can offer mechanistic insights into disease mechanisms and can complement future experimental efforts to complete the human interactome.
Drug combinations, offering increased therapeutic efficacy and reduced toxicity, play an important role in treating multiple complex diseases. Yet, our ability to identify and validate effective combinations is limited by a combinatorial explosion, driven by both the large number of drug pairs as well as dosage combinations. Here we propose a network-based methodology to identify clinically efficacious drug combinations for specific diseases. By quantifying the network-based relationship between drug targets and disease proteins in the human protein–protein interactome, we show the existence of six distinct classes of drug–drug–disease combinations. Relying on approved drug combinations for hypertension and cancer, we find that only one of the six classes correlates with therapeutic effects: if the targets of the drugs both hit disease module, but target separate neighborhoods. This finding allows us to identify and validate antihypertensive combinations, offering a generic, powerful network methodology to identify efficacious combination therapies in drug development.
Here we identify hundreds of new drug-disease associations for over 900 FDA-approved drugs by quantifying the network proximity of disease genes and drug targets in the human (protein–protein) interactome. We select four network-predicted associations to test their causal relationship using large healthcare databases with over 220 million patients and state-of-the-art pharmacoepidemiologic analyses. Using propensity score matching, two of four network-based predictions are validated in patient-level data: carbamazepine is associated with an increased risk of coronary artery disease (CAD) [hazard ratio (HR) 1.56, 95% confidence interval (CI) 1.12–2.18], and hydroxychloroquine is associated with a decreased risk of CAD (HR 0.76, 95% CI 0.59–0.97). In vitro experiments show that hydroxychloroquine attenuates pro-inflammatory cytokine-mediated activation in human aortic endothelial cells, supporting mechanistically its potential beneficial effect in CAD. In summary, we demonstrate that a unique integration of protein-protein interaction network proximity and large-scale patient-level longitudinal data complemented by mechanistic in vitro studies can facilitate drug repurposing.
Background Deep mining of healthcare data has provided maps of comorbidity relationships between diseases. In parallel, integrative multi-omics investigations have generated high-resolution molecular maps of putative relevance for understanding disease initiation and progression. Yet, it is unclear how to advance an observation of comorbidity relations (one disease to others) to a molecular understanding of the driver processes and associated biomarkers. Results Since Chronic Obstructive Pulmonary disease (COPD) has emerged as a central hub in temporal comorbidity networks, we developed a systematic integrative data-driven framework to identify shared disease-associated genes and pathways, as a proxy for the underlying generative mechanisms inducing comorbidity. We integrated records from approximately 13 M patients from the Medicare database with disease-gene maps that we derived from several resources including a semantic-derived knowledge-base. Using rank-based statistics we not only recovered known comorbidities but also discovered a novel association between COPD and digestive diseases. Furthermore, our analysis provides the first set of COPD co-morbidity candidate biomarkers, including IL15, TNF and JUP, and characterizes their association to aging and life-style conditions, such as smoking and physical activity. Conclusions The developed framework provides novel insights in COPD and especially COPD co-morbidity associated mechanisms. The methodology could be used to discover and decipher the molecular underpinning of other comorbidity relationships and furthermore, allow the identification of candidate co-morbidity biomarkers.
Historically, human diseases have been differentiated and categorized based on the organ system in which they primarily manifest. Recently, an alternative view is emerging that emphasizes that different diseases often have common underlying mechanisms and shared intermediate pathophenotypes, or endo(pheno)types. Within this framework, a specific disease’s expression is a consequence of the interplay between the relevant endophenotypes and their local, organ-based environment. Important examples of such endophenotypes are inflammation, fibrosis, and thrombosis and their essential roles in many developing diseases. In this study, we construct endophenotype network models and explore their relation to different diseases in general and to cardiovascular diseases in particular. We identify the local neighborhoods (module) within the interconnected map of molecular components, i.e., the subnetworks of the human interactome that represent the inflammasome, thrombosome, and fibrosome. We find that these neighborhoods are highly overlapping and significantly enriched with disease-associated genes. In particular they are also enriched with differentially expressed genes linked to cardiovascular disease (risk). Finally, using proteomic data, we explore how macrophage activation contributes to our understanding of inflammatory processes and responses. The results of our analysis show that inflammatory responses initiate from within the cross-talk of the three identified endophenotypic modules.
The protein-protein interaction (PPI) network is crucial for cellular information processing and decision-making. With suitable inputs, PPI networks drive the cells to diverse functional outcomes such as cell proliferation or cell death. Here, we characterize the structural controllability of a large directed human PPI network comprising 6,339 proteins and 34,813 interactions. This network allows us to classify proteins as "indispensable," "neutral," or "dispensable," which correlates to increasing, no effect, or decreasing the number of driver nodes in the network upon removal of that protein. We find that 21% of the proteins in the PPI network are indispensable. Interestingly, these indispensable proteins are the primary targets of disease-causing mutations, human viruses, and drugs, suggesting that altering a networks control property is critical for the transition between healthy and disease states. Furthermore, analyzing copy number alterations data from 1,547 cancer patients reveals that 56 genes that are frequently amplified or deleted in nine different cancers are indispensable. Among the 56 genes, 46 of them have not been previously associated with cancer. This suggests that controllability analysis is very useful in identifying novel disease genes and potential drug targets.
The increasing cost of drug development together with a significant drop in the number of new drug approvals raises the need for innovative approaches for target identification and efficacy prediction. Here, we take advantage of our increasing understanding of the network-based origins of diseases to introduce a drug-disease proximity measure that quantifies the interplay between drugs targets and diseases. By correcting for the known biases of the interactome, proximity helps us uncover the therapeutic effect of drugs, as well as to distinguish palliative from effective treatments. Our analysis of 238 drugs used in 78 diseases indicates that the therapeutic effect of drugs is localized in a small network neighborhood of the disease genes and highlights efficacy issues for drugs used in Parkinson and several inflammatory disorders. Finally, network-based proximity allows us to predict novel drug-disease associations that offer unprecedented opportunities for drug repurposing and the detection of adverse effects.
Genes carrying mutations associated with genetic diseases are present in all human cells; yet, clinical manifestations of genetic diseases are usually highly tissue-specific. Although some disease genes are expressed only in selected tissues, the expression patterns of disease genes alone cannot explain the observed tissue specificity of human diseases. Here we hypothesize that for a disease to manifest itself in a particular tissue, a whole functional subnetwork of genes (disease module) needs to be expressed in that tissue. Driven by this hypothesis, we conducted a systematic study of the expression patterns of disease genes within the human interactome. We find that genes expressed in a specific tissue tend to be localized in the same neighborhood of the interactome. By contrast, genes expressed in different tissues are segregated in distinct network neighborhoods. Most important, we show that it is the integrity and the completeness of the expression of the disease module that determines disease manifestation in selected tissues. This approach allows us to construct a disease-tissue network that confirms known and predicts unexpected disease-tissue associations.
According to the disease module hypothesis, the cellular components associated with a disease segregate in the same neighborhood of the human interactome, the map of biologically relevant molecular interactions. Yet, given the incompleteness of the interactome and the limited knowledge of disease-associated genes, it is not obvious if the available data have sufficient coverage to map out modules associated with each disease. Here we derive mathematical conditions for the identifiability of disease modules and show that the network-based location of each disease module determines its pathobiological relationship to other diseases. For example, diseases with overlapping network modules show significant coexpression patterns, symptom similarity, and comorbidity, whereas diseases residing in separated network neighborhoods are phenotypically distinct. These tools represent an interactome-based platform to predict molecular commonalities between phenotypically related diseases, even if they do not share primary disease genes.
Recent advances in genetics have spurred rapid progress towards the systematic identification of genes involved in complex diseases. Still, the detailed understanding of the molecular and physiological mechanisms through which these genes affect disease phenotypes remains a major challenge. Here, we identify the asthma disease module, i.e. the local neighborhood of the interactome whose perturbation is associated with asthma, and validate it for functional and pathophysiological relevance, using both computational and experimental approaches. We find that the asthma disease module is enriched with modest GWAS P-values against the background of random variation, and with differentially expressed genes from normal and asthmatic fibroblast cells treated with an asthma-specific drug. The asthma module also contains immune response mechanisms that are shared with other immune-related disease modules. Further, using diverse omics (genomics, gene-expression, drug response) data, we identify the GAB1 signaling pathway as an important novel modulator in asthma. The wiring diagram of the uncovered asthma module suggests a relatively close link between GAB1 and glucocorticoids (GCs), which we experimentally validate, observing an increase in the level of GAB1 after GC treatment in BEAS-2B bronchial epithelial cells. The siRNA knockdown of GAB1 in the BEAS-2B cell line resulted in a decrease in the NFkB level, suggesting a novel regulatory path of the pro-inflammatory factor NFkB by GAB1 in asthma.
Just as reference genome sequences revolutionized human genetics, reference maps of interactome networks will be critical to fully understand genotype-phenotype relationships. Here, we describe a systematic map of ∼14,000 high-quality human binary protein-protein interactions. At equal quality, this map is ∼30% larger than what is available from small-scale studies published in the literature in the last few decades. While currently available information is highly biased and only covers a relatively small portion of the proteome, our systematic map appears strikingly more homogeneous, revealing a “broader” human interactome network than currently appreciated. The map also uncovers significant interconnectivity between known and candidate cancer gene products, providing unbiased evidence for an expanded functional cancer landscape, while demonstrating how high-quality interactome models will help “connect the dots” of the genomic revolution.
In the post-genomic era, the elucidation of the relationship between the molecular origins of diseases and their resulting phenotypes is a crucial task for medical research. Here, we use a large-scale biomedical literature database to construct a symptom-based human disease network and investigate the connection between clinical manifestations of diseases and their underlying molecular interactions. We find that the symptom-based similarity of two diseases correlates strongly with the number of shared genetic associations and the extent to which their associated proteins interact. Moreover, the diversity of the clinical manifestations of a disease can be related to the connectivity patterns of the underlying protein interaction network. The comprehensive, high-quality map of disease–symptom relations can further be used as a resource helping to address important questions in the field of systems medicine, for example, the identification of unexpected associations between diseases, disease etiology research or drug design.
Genotypic differences greatly influence susceptibility and resistance to disease. Understanding genotype–phenotype relationships requires that phenotypes be viewed as manifestations of network properties, rather than simply as the result of individual genomic variations. Genome sequencing efforts have identified numerous germline mutations, and large numbers of somatic genomic alterations, associated with a predisposition to cancer. However, it remains difficult to distinguish background, or ‘passenger’, cancer mutations from causal, or ‘driver’, mutations in these data sets. Human viruses intrinsically depend on their host cell during the course of infection and can elicit pathological phenotypes similar to those arising from mutations. Here we test the hypothesis that genomic variations and tumour viruses may cause cancer through related mechanisms, by systematically examining host interactome and transcriptome network perturbations caused by DNA tumour virus proteins. The resulting integrated viral perturbation data reflects rewiring of the host cell networks, and highlights pathways, such as Notch signalling and apoptosis, that go awry in cancer. We show that systematic analyses of host targets of viral proteins can identify cancer genes with a success rate on a par with their identification through functional genomics and large-scale cataloguing of tumour mutations. Together, these complementary approaches increase the specificity of cancer gene identification. Combining systems-level studies of pathogen-encoded gene products with genomic approaches will facilitate the prioritization of cancer causing driver genes to advance the understanding of the genetic basis of human cancer.
Contemporary views of human disease are based on simple correlation between clinical syndromes and pathological analysis dating from the late 19th century. Although this approach to disease diagnosis, prognosis, and treatment has served the medical establishment and society well for many years, it has serious shortcomings for the modern era of the genomic medicine that stem from its reliance on reductionist principles of experimentation and analysis. Quantitative, holistic systems biology applied to human disease offers a unique approach for diagnosing established disease, defining disease predilection, and developing individualized (personalized) treatment strategies that can take full advantage of modern molecular pathobiology and the comprehensive data sets that are rapidly becoming available for populations and individuals. In this way, systems pathobiology offers the promise of redefining our approach to disease and the field of medicine.
Complex biological systems and cellular networks may underlie most genotype to phenotype relationships. Here, we review basic concepts in network biology, discussing different types of interactome networks and the insights that can come from analyzing them. We elaborate on why interactome networks are important to consider in biology, how they can be mapped and integratedwith each other, what global properties are starting to emerge from interactome network models, and how these properties may relate to human disease.
By Albert-László Barabási, Natali Gulbahce & Joseph Loscalzo
Published: 17 December 2010
The use of networks to integrate different genetic, proteomic, and metabolic datasets has been proposed as a viable path toward elucidating the origins of specific diseases. Here we introduce a new phenotypic database summarizing correlations obtained from the disease history of more than 30 million patients in a Phenotypic Disease Network (PDN). We present evidence that the structure of the PDN is relevant to the understanding of illness progression by showing that (1) patients develop diseases close in the network to those they already have; (2) the progression of disease along the links of the network is different for patients of different genders and ethnicities; (3) patients diagnosed with diseases which are more highly connected in the PDN tend to die sooner than those affected by less connected diseases; and (4) diseases that tend to be preceded by others in the PDN tend to be more connected than diseases that precede other illnesses, and are associated with higher degrees of mortality. Our findings show that disease progression can be represented and studied using network methods, offering the potential to enhance our understanding of the origin and evolution of human diseases. The dataset introduced here, released concurrently with this publication, represents the largest relational phenotypic resource publicly available to the research community.
The impact of disease-causing defects is often not limited to the products of a mutated gene but, thanks to interactions between the molecular components, may also affect other cellular functions, resulting in potential comorbidity effects. By combining information on cellular interactions, disease--gene associations, and population-level disease patterns extracted from Medicare data, we find statistically significant correlations between the underlying structure of cellular networks and disease comorbidity patterns in the human population. Our results indicate that such a combination of population-level data and cellular network information could help build novel hypotheses about disease mechanisms.
Most diseases are the consequence of the breakdown of cellular processes, but the relationships among genetic/epigenetic defects, the molecular interaction networks underlying them, and the disease phenotypes remain poorly understood. To gain insights into such relationships, here we constructed a bipartite human disease association network in which nodes are diseases and two diseases are linked if mutated enzymes associated with them catalyze adjacent metabolic reactions. We find that connected disease pairs display higher correlated reaction flux rate, corresponding enzyme-encoding gene coexpression, and higher comorbidity than those that have no metabolic link between them. Furthermore, the more connected a disease is to other diseases, the higher is its prevalence and associated mortality rate. The network topology-based approach also helps to
The global set of relationships between protein targets of all drugs and all disease-gene products in the human protein–protein interaction or ‘interactome’ network remains uncharacterized. We built a bipartite graph composed of US Food and Drug Administration–approved drugs and proteins linked by drug–target binary associations. The resultingnetwork connects most drugs into a highly interlinked giant component, with strong local clustering of drugs of similar types according to Anatomical Therapeutic Chemical classification. Topological analyses of this network quantitatively showed an overabundance of ‘follow-on’ drugs, that is, drugs that target already targeted proteins. By including drugs currently under investigation, we identified a trend toward more functionally diverse targets improving polypharmacology. To analyze the relationships between drug targets and disease-gene products, we measured the shortest distance between both sets of proteins in current models of the human interactome network. Significant differences in distance were found between etiological and palliative drugs. A recent trend toward more rational drug design was observed.
Over the past decades, the diversity of areas explored by physicists has exploded, encompassing new topics from biophysics and chemical physics to network science. However, it is unclear how these new subfields emerged from the traditional subject areas and how physicists explore them. To map out the evolution of physics subfields, here, we take an intellectual census of physics by studying physicists’ careers. We use a large-scale publication data set, identify the subfields of 135,877 physicists and quantify their heterogeneous birth, growth and migration patterns among research areas. We find that the majority of physicists began their careers in only three subfields, branching out to other areas at later career stages, with different rates and transition times. Furthermore, we analyse the productivity, impact and team sizes across different subfields, finding drastic changes attributable to the recent rise in large-scale collaborations. This detailed, longitudinal census of physics can inform resource allocation policies and provide students, editors and scientists with a broader view of the field’s internal dynamics.
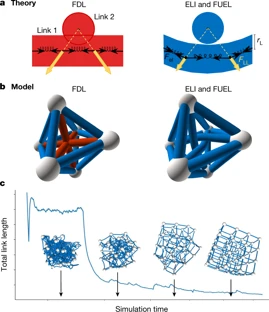
In many physical networks, including neurons in the brain three-dimensional integrated circuits and underground hyphal networks, the nodes and links are physical objects that cannot intersect or overlap with each other. To take this into account, non-crossing conditions can be imposed to constrain the geometry of networks, which consequently affects how they form, evolve and function. However, these constraints are not included in the theoretical frameworks that are currently used to characterize real networks. Most tools for laying out networks are variants of the force-directed layout algorithm—which assumes dimensionless nodes and links—and are therefore unable to reveal the geometry of densely packed physical networks. Here we develop a modelling framework that accounts for the physical sizes of nodes and links, allowing us to explore how non-crossing conditions affect the geometry of a network. For small link thicknesses, we observe a weakly interacting regime in which link crossings are avoided via local link rearrangements, without altering the overall geometry of the layout compared to the force-directed layout. Once the link thickness exceeds a threshold, a strongly interacting regime emerges in which multiple geometric quantities, such as the total link length and the link curvature, scale with the link thickness. We show that the crossover between the two regimes is driven by the non-crossing condition, which allows us to derive the transition point analytically and show that networks with large numbers of nodes will ultimately exist in the strongly interacting regime. We also find that networks in the weakly interacting regime display a solid-like response to stress, whereas in the strongly interacting regime they behave in a gel-like fashion. Networks in the weakly interacting regime are amenable to 3D printing and so can be used to visualize network geometry, and the strongly interacting regime provides insights into the scaling of the sizes of densely packed mammalian brains.
High-throughput technologies, offering an unprecedented wealth of quantitative data underlying the makeup of living systems, are changing biology. Notably, the systematic mapping of the relationships between biochemical entities has fueled the rapid development of network biology, offering a suitable framework to describe disease phenotypes and predict potential drug targets. However, our ability to develop accurate dynamical models remains limited, due in part to the limited knowledge of the kinetic parameters underlying these interactions. Here, we explore the degree to which we can make reasonably accurate predictions in the absence of the kinetic parameters. We find that simple dynamically agnostic models are sufficient to recover the strength and sign of the biochemical perturbation patterns observed in 87 biological models for which the underlying kinetics are known. Surprisingly, a simple distance-based model achieves 65% accuracy. We show that this predictive power is robust to topological and kinetic parameter perturbations, and we identify key network properties that can increase up to 80% the recovery rate of the true perturbation patterns. We validate our approach using experimental data on the chemotactic pathway in bacteria, finding that a network model of perturbation spreading predicts with ∼80% accuracy the directionality of gene expression and phenotype changes in knock-out and overproduction experiments. These findings show that the steady advances in mapping out the topology of biochemical interaction networks opens avenues for accurate perturbation spread modeling, with direct implications for medicine and drug development.
Most networked systems of scientific interest are characterized by temporal links, meaning the network’s structure changes over time. Link temporality has been shown to hinder many dynamical processes, from information spreading to accessibility, by disrupting network paths. Considering the ubiquity of temporal networks in nature, we ask: Are there any advantages of the networks’ temporality? We use an analytical framework to show that temporal networks can, compared to their static counterparts, reach controllability faster, demand orders of magnitude less control energy, and have control trajectories, that are considerably more compact than those characterizing static networks. Thus, temporality ensures a degree of flexibility that would be unattainable in static networks, enhancing our ability to control them.
A physicist probes a phenomenon seen in cells, cities, and almost everything in between.
Pinpointing the nodes whose removal most effectively disrupts a network has become a lot easier with the development of an efficient algorithm. Potential applications might include cybersecurity and disease control. See Letter p.65, by F. Morone and H. A. Makse (Supplementary 1).
Professor Barabási's talk described how the tools of network science can help understand the Web's structure, development and weaknesses. The Web is an information network, in which the nodes are documents (at the time of writing over one trillion of them), connected by links. Other well-known network structures include the Internet, a physical network where the nodes are routers and the links are physical connections, and organizations, where the nodes are people and the links represent communications.
The concept of preferential attachment is behind the hubs and power laws seen in many networks. New results fuel an old debate about its origin, and beg the question of whether it is based on randomness or optimization.
We are surrounded by complex systems, from cells made of thousands of molecules to society, a collection of billions of interacting individuals. These systems display signatures of order and self-organization. Understanding and quantifying this complexity is a grand challenge for science. Kinetic theory, developed at the end of the 19th century, shows that the measurable properties of gases, from pressure to temperature, can be reduced to the random motion of atoms and molecules. In the 1960s and 1970s, researchers developed systematic approaches to quantifying the transition from disorder to order in material systems such as magnets and liquids. Chaos theory dominated the quest to understand complex behavior in the 1980s with the message that unpredictable behavior can emerge from the nonlinear interactions of a few components. The 1990s was the decade of fractals, quantifying the geometry of patterns emerging in self-organized systems, from leaves to snowflakes.
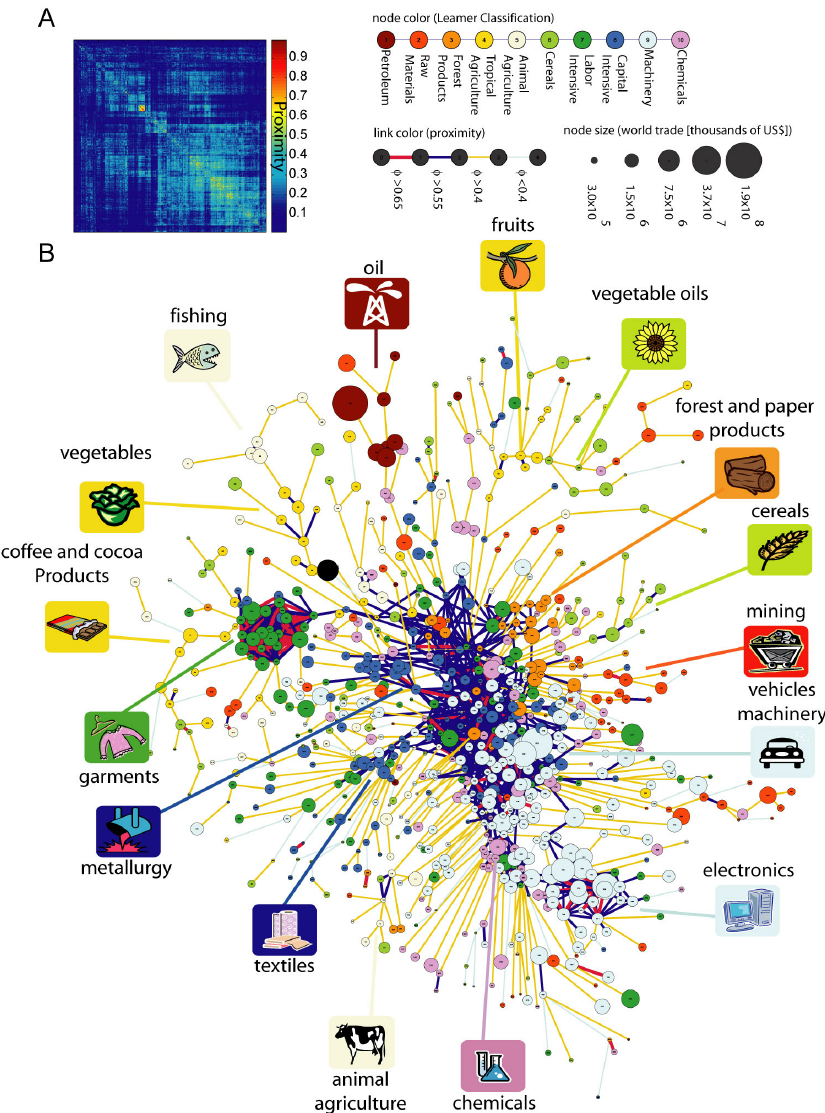
Economies grow by upgrading the products they produce and export. The technology, capital, institutions, and skills needed to make newer products are more easily adapted from some products than from others. Here, we study this network of relatedness between products, or “product space,” finding that more-sophisticated products are located in a densely connected core whereas lesssophisticated products occupy a less-connected periphery. Empirically, countries move through the product space by developing goods close to those they currently produce. Most countries can reach the core only by traversing empirically infrequent distances, which may help explain why poor countries have trouble developing more competitive exports and fail to converge to the income levels of rich countries.
We construct a connected network of 3.9 million nodes from mobile phone call records, which can be regarded as a proxy for the underlying human communication network at the societal level. We assign two weights on each edge to reflect the strength of social interaction, which are the aggregate call duration and the cumulative number of calls placed between the individuals over a period of 18 weeks. We present a detailed analysis of this weighted network by examining its degree, strength, and weight distributions, as well as its topological assortativity and weighted assortativity, clustering and weighted clustering, together with correlations between these quantities.We give an account of motif intensity and coherence distributions and compare them to a randomized reference system.We also use the concept of link overlap to measure the number of common neighbours any two adjacent nodes have, which serves as a useful local measure for identifying the interconnectedness of communities. We report a positive correlation between the overlap and weight of a link, thus providing strong quantitative evidence for the weak ties hypothesis, a central concept in social network analysis. The percolation properties of the network are found to depend on the type and order of removed links, and they can help understand how the local structure of the network manifests itself at the global level.We hope that our results will contribute to modelling weighted large-scale social networks, and believe that the systematic approach followed here can be adopted to study other weighted networks.
Electronic databases, from phone to e-mails logs, currently provide detailed records of human communication patterns, offering novel avenues to map and explore the structure of social and communication networks. Here we examine the communication patterns of millions of mobile phone users, allowing us to simultaneously study the local and the global structure of a society-wide communication network. We observe a coupling between interaction strengths and the network's local structure, with the counterintuitive consequence that social networks are robust to the removal of the strong ties but fall apart after a phase transition if the weak ties are removed. We show that this coupling significantly slows the diffusion process, resulting in dynamic trapping of information in communities and find that, when it comes to information diffusion, weak and strong ties are both simultaneously ineffective.
Our focus is on networks capturing the collaboration between scientists and the calls between mobile phone users. We find that large groups persist for longer if they are capable of dynamically altering their membership, suggesting that an ability to change the group composition results in better adaptability. The behaviour of small groups displays the opposite tendency—the condition for stability is that their composition remains unchanged. We also show that knowledge of the time commitment of members to a given community can be used for estimating the community’s lifetime. These findings offer insight into the fundamental differences between the dynamics of small groups and large institutions.
While current studies on complex networks focus on systems that change relatively slowly in time, the structure of the most visited regions of the web is altered at the time scale from hours to days. Here we investigate the dynamics of visitation of a major news portal, representing the prototype for such a rapidly evolving network. The nodes of the network can be classified into stable nodes, which form the timeindependent skeleton of the portal, and news documents. The visitations of the two node classes are markedly different, the skeleton acquiring visits at a constant rate, while a news document’s visitation peaks after a few hours. We find that the visitation pattern of a news document decays as a power law, in contrast with the exponential prediction provided by simple models of site visitation. This is rooted in the inhomogeneous nature of the browsing pattern characterizing individual users: the time interval between consecutive visits by the same user to the site follows a power-law distribution, in contrast to the exponential expected for Poisson processes. We show that the exponent characterizing the individual user’s browsing patterns determines the power-law decay in a document’s visitation. Finally, our results document the fleeting quality of news and events: while fifteen minutes of fame is still an exaggeration in the online media, we find that access to most news items significantly decays after 36 hours of posting.
The science of networks is experiencing a boom. But despite the necessary multidisciplinary approach to tackle the theory of complexity, scientists remain largely compartmentalized in their separate disciplines. Can they find a common voice?
A key aim of postgenomic biomedical research is to systematically catalogue all molecules and their interactions within a living cell. There is a clear need to understand how these molecules and the interactions between them determine the function of this enormously complex machinery, both in isolation and when surrounded by other cells. Rapid advances in network biology indicate that cellular networks are governed by universal laws and offer a new conceptual framework that could potentially revolutionize our view of biology and disease pathologies in the twenty-first century.
Scientists have recently discovered that various complex systems have an underlying architecture governed by shared organizing principies. This insight has important implications for a host of applications, from drug development to Internet security.
Network generators that capture the Internet’s large-scale topology are crucial for the development of efficient routing protocols and modeling Internet traffic. Our ability to design realistic generators is limited by the incomplete understanding of the fundamental driving forces that affect the Internet’s evolution. By combining several independent databases capturing the time evolution, topology, and physical layout of the Internet, we identify the universal mechanisms that shape the Internet’s router and autonomous system level topology. We find that the physical layout of nodes form a fractal set, determined by population density patterns around the globe. The placement of links is driven by competition between preferential attachment and linear distance dependence, a marked departure from the currently used exponential laws. The universal parameters that we extract significantly restrict the class of potentially correct Internet models and indicate that the networks created by all available topology generators are fundamentally different from the current Internet.
The internet appears to have taken on a life of its own ever since the National Science Foundation in the US gave up stewardship of the network in 1995. New lines and routers are added continually by thousands of companies, none of which require permission from anybody to do so, and none of which are obliged to report their activity. This uncontrolled and decentralized growth has turned network designers into scientific explorers. All previous Internet-related research concentrated on designing better protocols and faster components. More recently, an increasing number of scientists have begun to ask an unexpected question: what exactly did we create?
The most highly connected proteins in the cell are the most important for its survival. Proteins are traditionally identified on the basis of their individual actions as catalysts, signalling molecules, or building blocks in cells and microorganisms. But our post-genomic view is expanding the protein’s role into an element in a network of protein–protein interactions as well, in which it has a contextual or cellular function within functional modules1,2. Here we provide quantitative support for this idea by demonstrating that the phenotypic consequence of a single gene deletion in the yeast Saccharomyces cerevisiae is affected to a large extent by the topological position of its protein product in the complex hierarchical web of molecular interactions.
Here we present a systematic comparative mathematical analysis of the metabolic networks of 43 organisms representing all three domains of life.We show that, despite significant variation in their individual constituents and pathways, these metabolic networks have the same topological scaling properties and show striking similarities to the inherent organization of complex non-biological systems. This may indicate that metabolic organization is not only identical for all living organisms, but also complies with the design principles of robust and error-tolerant scale-free networks, and may represent a common blueprint for the large-scale organization of interactions among all cellular constituents.
Here we demonstrate that error tolerance is not shared by all redundant systems: it is displayed only by a class of inhomogeneouslywired networks,called scale-free networks, which include theWorld-WideWeb, the Internet, social networks and cells. We find that such networks display an unexpected degree of robustness, the ability of their nodes to communicate being unaffected even by unrealistically high failure rates.However, error tolerance comes at a high price in that these networks are extremely vulnerable to attacks (that is, to the selection and removal of a few nodes that play a vital role in maintaining the network’s connectivity). Such error tolerance and attack vulnerability are generic properties of communication networks.
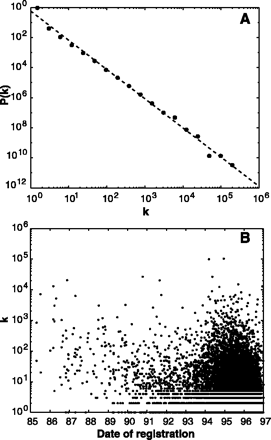
The world-wide web forms a large directed graph, whose vertices are documents and edges are links pointing from one document to another. Here we demonstrate that despite its apparent random character, the topology of this graph has a number of universal scale-free characteristics. We introduce a model that leads to a scale-free network, capturing in a minimal fashion the self-organization processes governing the world-wide web.
Barabasi and Albert propose an improved version of the Erdos-Renyi theory of random networks to account for the scaling properties of a number of systems, including the link structure of the World Wide Web (WWW). The theory they present, however, is inconsistent with empirically observed properties of the Web link structure.
Systems as diverse as genetic networks or the World Wide Web are best described as networks with complex topology. A common property of many large networks is that the vertex connectivities follow a scale-free power-law distribution. This feature was found to be a consequence of two generic mechanisms: (i) networks expand continuously by the addition of new vertices, and (ii) new vertices attach preferentially to sites that are already well connected. A model based on these two ingredients reproduces the observed stationary scale-free distributions, which indicates that the development of large networks is governed by robust self-organizing phenomena that go beyond the particulars of the individual systems.
Despite its increasing role in communication, the World-Wide Web remains uncontrolled: any individual or institution can create a website with any number of documents and links. This unregulated growth leads to a huge and complex web, which becomes a large directed graph whose vertices are documents and whose edges are links (URLs) that point from one document to another. The topology of this graph determines the web’s connectivity and consequently how effectively we can locate information on it.
News
The causes of complex diseases can be identified by representing them in the form of mathematically produced networks. This method was used to find bacteria that drive atopic dermatitis, for example.
Complex disease definitions often represent descriptive umbrella terms of symptoms rather than mechanistic
entities. A new study shows how network-based approaches can help identify the mechanisms that link genes,
cells, tissues and organs in cardiovascular diseases.
Second International Conference on Network Medicine and Big Data
April 12-13, 2021
Virtual/Boston, MA
Hosted by:
Brigham and Women’s Hospital and Harvard Medical School