Taking Census of Physics
Over the past decades, the diversity of areas explored by physicists has exploded, encompassing new topics from biophysics and chemical physics to network science. However, it is unclear how these new subfields emerged from the traditional subject areas and how physicists explore them. To map out the evolution of physics subfields, here, we take an intellectual census of physics by studying physicists’ careers. We use a large-scale publication data set, identify the subfields of 135,877 physicists and quantify their heterogeneous birth, growth and migration patterns among research areas. We find that the majority of physicists began their careers in only three subfields, branching out to other areas at later career stages, with different rates and transition times. Furthermore, we analyse the productivity, impact and team sizes across different subfields, finding drastic changes attributable to the recent rise in large-scale collaborations. This detailed, longitudinal census of physics can inform resource allocation policies and provide students, editors and scientists with a broader view of the field’s internal dynamics.
A Structural Transition in Physical Networks
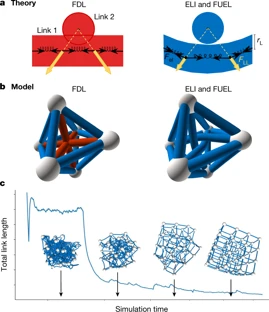
In many physical networks, including neurons in the brain three-dimensional integrated circuits and underground hyphal networks, the nodes and links are physical objects that cannot intersect or overlap with each other. To take this into account, non-crossing conditions can be imposed to constrain the geometry of networks, which consequently affects how they form, evolve and function. However, these constraints are not included in the theoretical frameworks that are currently used to characterize real networks. Most tools for laying out networks are variants of the force-directed layout algorithm—which assumes dimensionless nodes and links—and are therefore unable to reveal the geometry of densely packed physical networks. Here we develop a modelling framework that accounts for the physical sizes of nodes and links, allowing us to explore how non-crossing conditions affect the geometry of a network. For small link thicknesses, we observe a weakly interacting regime in which link crossings are avoided via local link rearrangements, without altering the overall geometry of the layout compared to the force-directed layout. Once the link thickness exceeds a threshold, a strongly interacting regime emerges in which multiple geometric quantities, such as the total link length and the link curvature, scale with the link thickness. We show that the crossover between the two regimes is driven by the non-crossing condition, which allows us to derive the transition point analytically and show that networks with large numbers of nodes will ultimately exist in the strongly interacting regime. We also find that networks in the weakly interacting regime display a solid-like response to stress, whereas in the strongly interacting regime they behave in a gel-like fashion. Networks in the weakly interacting regime are amenable to 3D printing and so can be used to visualize network geometry, and the strongly interacting regime provides insights into the scaling of the sizes of densely packed mammalian brains.
Predicting Perturbation Patterns from the Topology of Biological Networks
High-throughput technologies, offering an unprecedented wealth of quantitative data underlying the makeup of living systems, are changing biology. Notably, the systematic mapping of the relationships between biochemical entities has fueled the rapid development of network biology, offering a suitable framework to describe disease phenotypes and predict potential drug targets. However, our ability to develop accurate dynamical models remains limited, due in part to the limited knowledge of the kinetic parameters underlying these interactions. Here, we explore the degree to which we can make reasonably accurate predictions in the absence of the kinetic parameters. We find that simple dynamically agnostic models are sufficient to recover the strength and sign of the biochemical perturbation patterns observed in 87 biological models for which the underlying kinetics are known. Surprisingly, a simple distance-based model achieves 65% accuracy. We show that this predictive power is robust to topological and kinetic parameter perturbations, and we identify key network properties that can increase up to 80% the recovery rate of the true perturbation patterns. We validate our approach using experimental data on the chemotactic pathway in bacteria, finding that a network model of perturbation spreading predicts with ∼80% accuracy the directionality of gene expression and phenotype changes in knock-out and overproduction experiments. These findings show that the steady advances in mapping out the topology of biochemical interaction networks opens avenues for accurate perturbation spread modeling, with direct implications for medicine and drug development.
The Fundamental Advantages of Temporal Networks
Most networked systems of scientific interest are characterized by temporal links, meaning the network’s structure changes over time. Link temporality has been shown to hinder many dynamical processes, from information spreading to accessibility, by disrupting network paths. Considering the ubiquity of temporal networks in nature, we ask: Are there any advantages of the networks’ temporality? We use an analytical framework to show that temporal networks can, compared to their static counterparts, reach controllability faster, demand orders of magnitude less control energy, and have control trajectories, that are considerably more compact than those characterizing static networks. Thus, temporality ensures a degree of flexibility that would be unattainable in static networks, enhancing our ability to control them.
The Elegant Law that Governs Us All
A physicist probes a phenomenon seen in cells, cities, and almost everything in between.
Destruction perfected
Pinpointing the nodes whose removal most effectively disrupts a network has become a lot easier with the development of an efficient algorithm. Potential applications might include cybersecurity and disease control. See Letter p.65, by F. Morone and H. A. Makse (Supplementary 1).
Network Science
Professor Barabási's talk described how the tools of network science can help understand the Web's structure, development and weaknesses. The Web is an information network, in which the nodes are documents (at the time of writing over one trillion of them), connected by links. Other well-known network structures include the Internet, a physical network where the nodes are routers and the links are physical connections, and organizations, where the nodes are people and the links represent communications.
Network science: Luck or reason
The concept of preferential attachment is behind the hubs and power laws seen in many networks. New results fuel an old debate about its origin, and beg the question of whether it is based on randomness or optimization.
The architecture of complexity
We are surrounded by complex systems, from cells made of thousands of molecules to society, a collection of billions of interacting individuals. These systems display signatures of order and self-organization. Understanding and quantifying this complexity is a grand challenge for science. Kinetic theory, developed at the end of the 19th century, shows that the measurable properties of gases, from pressure to temperature, can be reduced to the random motion of atoms and molecules. In the 1960s and 1970s, researchers developed systematic approaches to quantifying the transition from disorder to order in material systems such as magnets and liquids. Chaos theory dominated the quest to understand complex behavior in the 1980s with the message that unpredictable behavior can emerge from the nonlinear interactions of a few components. The 1990s was the decade of fractals, quantifying the geometry of patterns emerging in self-organized systems, from leaves to snowflakes.
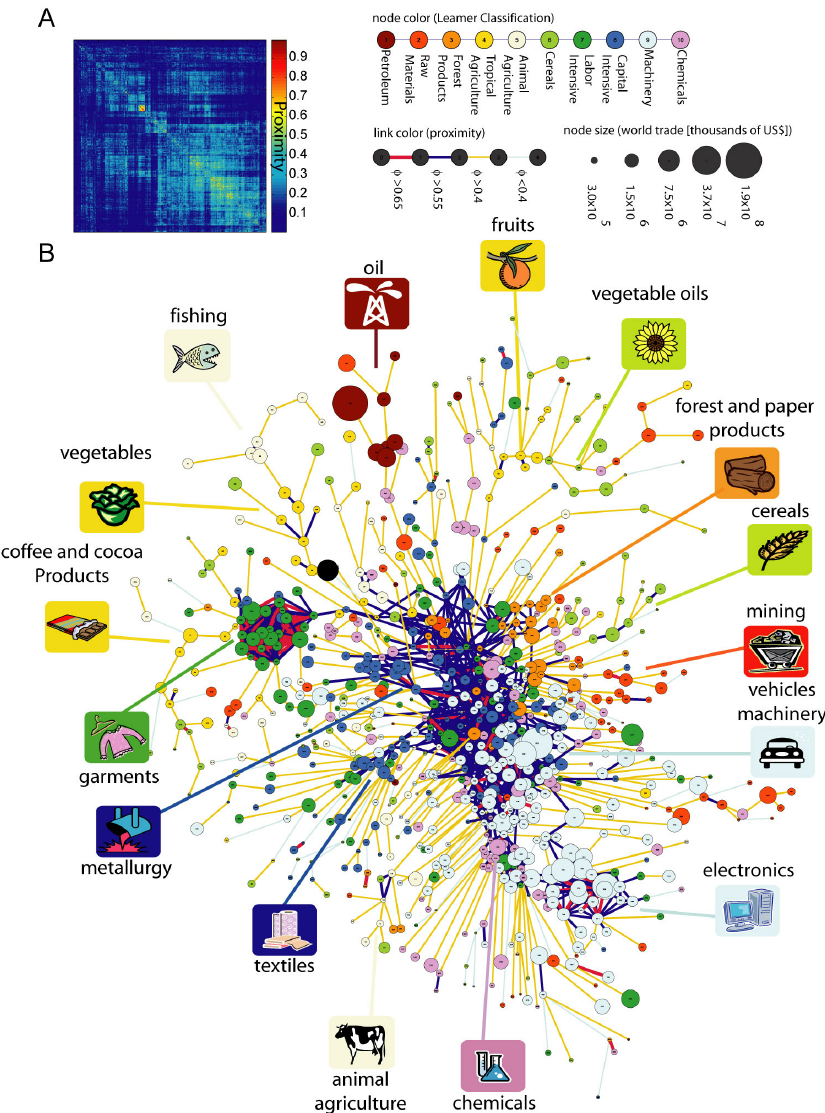
The product space conditions the development of nations
Economies grow by upgrading the products they produce and export. The technology, capital, institutions, and skills needed to make newer products are more easily adapted from some products than from others. Here, we study this network of relatedness between products, or “product space,” finding that more-sophisticated products are located in a densely connected core whereas lesssophisticated products occupy a less-connected periphery. Empirically, countries move through the product space by developing goods close to those they currently produce. Most countries can reach the core only by traversing empirically infrequent distances, which may help explain why poor countries have trouble developing more competitive exports and fail to converge to the income levels of rich countries.
Analysis of a large-scale weighted network of one-to-one human communication
We construct a connected network of 3.9 million nodes from mobile phone call records, which can be regarded as a proxy for the underlying human communication network at the societal level. We assign two weights on each edge to reflect the strength of social interaction, which are the aggregate call duration and the cumulative number of calls placed between the individuals over a period of 18 weeks. We present a detailed analysis of this weighted network by examining its degree, strength, and weight distributions, as well as its topological assortativity and weighted assortativity, clustering and weighted clustering, together with correlations between these quantities.We give an account of motif intensity and coherence distributions and compare them to a randomized reference system.We also use the concept of link overlap to measure the number of common neighbours any two adjacent nodes have, which serves as a useful local measure for identifying the interconnectedness of communities. We report a positive correlation between the overlap and weight of a link, thus providing strong quantitative evidence for the weak ties hypothesis, a central concept in social network analysis. The percolation properties of the network are found to depend on the type and order of removed links, and they can help understand how the local structure of the network manifests itself at the global level.We hope that our results will contribute to modelling weighted large-scale social networks, and believe that the systematic approach followed here can be adopted to study other weighted networks.
Structure and tie strengths in mobile communication networks
Electronic databases, from phone to e-mails logs, currently provide detailed records of human communication patterns, offering novel avenues to map and explore the structure of social and communication networks. Here we examine the communication patterns of millions of mobile phone users, allowing us to simultaneously study the local and the global structure of a society-wide communication network. We observe a coupling between interaction strengths and the network's local structure, with the counterintuitive consequence that social networks are robust to the removal of the strong ties but fall apart after a phase transition if the weak ties are removed. We show that this coupling significantly slows the diffusion process, resulting in dynamic trapping of information in communities and find that, when it comes to information diffusion, weak and strong ties are both simultaneously ineffective.
Quantifying social group evolution
Our focus is on networks capturing the collaboration between scientists and the calls between mobile phone users. We find that large groups persist for longer if they are capable of dynamically altering their membership, suggesting that an ability to change the group composition results in better adaptability. The behaviour of small groups displays the opposite tendency—the condition for stability is that their composition remains unchanged. We also show that knowledge of the time commitment of members to a given community can be used for estimating the community’s lifetime. These findings offer insight into the fundamental differences between the dynamics of small groups and large institutions.
Dynamics of information access on the web
While current studies on complex networks focus on systems that change relatively slowly in time, the structure of the most visited regions of the web is altered at the time scale from hours to days. Here we investigate the dynamics of visitation of a major news portal, representing the prototype for such a rapidly evolving network. The nodes of the network can be classified into stable nodes, which form the timeindependent skeleton of the portal, and news documents. The visitations of the two node classes are markedly different, the skeleton acquiring visits at a constant rate, while a news document’s visitation peaks after a few hours. We find that the visitation pattern of a news document decays as a power law, in contrast with the exponential prediction provided by simple models of site visitation. This is rooted in the inhomogeneous nature of the browsing pattern characterizing individual users: the time interval between consecutive visits by the same user to the site follows a power-law distribution, in contrast to the exponential expected for Poisson processes. We show that the exponent characterizing the individual user’s browsing patterns determines the power-law decay in a document’s visitation. Finally, our results document the fleeting quality of news and events: while fifteen minutes of fame is still an exaggeration in the online media, we find that access to most news items significantly decays after 36 hours of posting.
Taming complexity
The science of networks is experiencing a boom. But despite the necessary multidisciplinary approach to tackle the theory of complexity, scientists remain largely compartmentalized in their separate disciplines. Can they find a common voice?
Network biology: understanding the cell’s functional organization
A key aim of postgenomic biomedical research is to systematically catalogue all molecules and their interactions within a living cell. There is a clear need to understand how these molecules and the interactions between them determine the function of this enormously complex machinery, both in isolation and when surrounded by other cells. Rapid advances in network biology indicate that cellular networks are governed by universal laws and offer a new conceptual framework that could potentially revolutionize our view of biology and disease pathologies in the twenty-first century.
Scale-Free Networks
Scientists have recently discovered that various complex systems have an underlying architecture governed by shared organizing principies. This insight has important implications for a host of applications, from drug development to Internet security.
Modeling the internet’s large-scale topology
Network generators that capture the Internet’s large-scale topology are crucial for the development of efficient routing protocols and modeling Internet traffic. Our ability to design realistic generators is limited by the incomplete understanding of the fundamental driving forces that affect the Internet’s evolution. By combining several independent databases capturing the time evolution, topology, and physical layout of the Internet, we identify the universal mechanisms that shape the Internet’s router and autonomous system level topology. We find that the physical layout of nodes form a fractal set, determined by population density patterns around the globe. The placement of links is driven by competition between preferential attachment and linear distance dependence, a marked departure from the currently used exponential laws. The universal parameters that we extract significantly restrict the class of potentially correct Internet models and indicate that the networks created by all available topology generators are fundamentally different from the current Internet.
The physics of the web
The internet appears to have taken on a life of its own ever since the National Science Foundation in the US gave up stewardship of the network in 1995. New lines and routers are added continually by thousands of companies, none of which require permission from anybody to do so, and none of which are obliged to report their activity. This uncontrolled and decentralized growth has turned network designers into scientific explorers. All previous Internet-related research concentrated on designing better protocols and faster components. More recently, an increasing number of scientists have begun to ask an unexpected question: what exactly did we create?
Lethality and centrality in protein networks
The most highly connected proteins in the cell are the most important for its survival. Proteins are traditionally identified on the basis of their individual actions as catalysts, signalling molecules, or building blocks in cells and microorganisms. But our post-genomic view is expanding the protein’s role into an element in a network of protein–protein interactions as well, in which it has a contextual or cellular function within functional modules1,2. Here we provide quantitative support for this idea by demonstrating that the phenotypic consequence of a single gene deletion in the yeast Saccharomyces cerevisiae is affected to a large extent by the topological position of its protein product in the complex hierarchical web of molecular interactions.