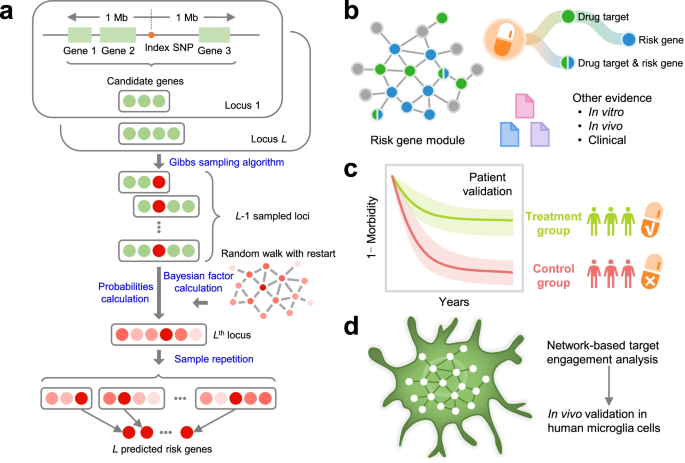
Artificial intelligence framework identifies candidate targets for drug repurposing in Alzheimer’s disease
Genome-wide association studies (GWAS) have identified numerous susceptibility loci for Alzheimer’s disease (AD). However, utilizing GWAS and multi-omics data to identify high-confidence AD risk genes (ARGs) and druggable targets that can guide development of new therapeutics for patients suffering from AD has heretofore not been successful.
Endophenotype-based in silico network medicine discovery combined with insurance record data mining identifies sildenafil as a candidate drug for Alzheimer’s disease
We developed an endophenotype disease module-based methodology for Alzheimer’s disease (AD) drug repurposing and identified sildenafil as a potential disease risk modifier. Based on retrospective case–control pharmacoepidemiologic analyses of insurance claims data for 7.23 million individuals, we found that sildenafil usage was significantly associated with a 69% reduced risk of AD (hazard ratio 0.31, 95% confidence interval 0.25–0.39, P < 1.0 × 10–8). Propensity score-stratified analyses confirmed that sildenafil is significantly associated with a decreased risk of AD across all four drug cohorts tested (diltiazem, glimepiride, losartan and metformin) after adjusting for age, sex, race and disease comorbidities. We also found that sildenafil increases neurite growth and decreases phospho-tau expression in neuron models derived from induced pluripotent stem cells from patients with AD, supporting mechanistically its potential beneficial effect in AD. The association between sildenafil use and decreased incidence of AD does not establish causality, which will require a randomized controlled trial.
Network medicine for disease module identification and drug repurposing with the NeDRex platform
Traditional drug discovery faces a severe efficacy crisis. Repurposing of registered drugs provides an alternative with lower costs and faster drug development timelines. However, the data necessary for the identification of disease modules, i.e. pathways and sub-networks describing the mechanisms of complex diseases which contain potential drug targets, are scattered across independent databases. Moreover, existing studies are limited to predictions for specific diseases or non-translational algorithmic approaches. There is an unmet need for adaptable tools allowing biomedical researchers to employ network-based drug repurposing approaches for their individual use cases. We close this gap with NeDRex, an integrative and interactive platform for network-based drug repurposing and disease module discovery. NeDRex integrates ten different data sources covering genes, drugs, drug targets, disease annotations, and their relationships. NeDRex allows for constructing heterogeneous biological networks, mining them for disease modules, prioritizing drugs targeting disease mechanisms, and statistical validation. We demonstrate the utility of NeDRex in five specific use-cases.
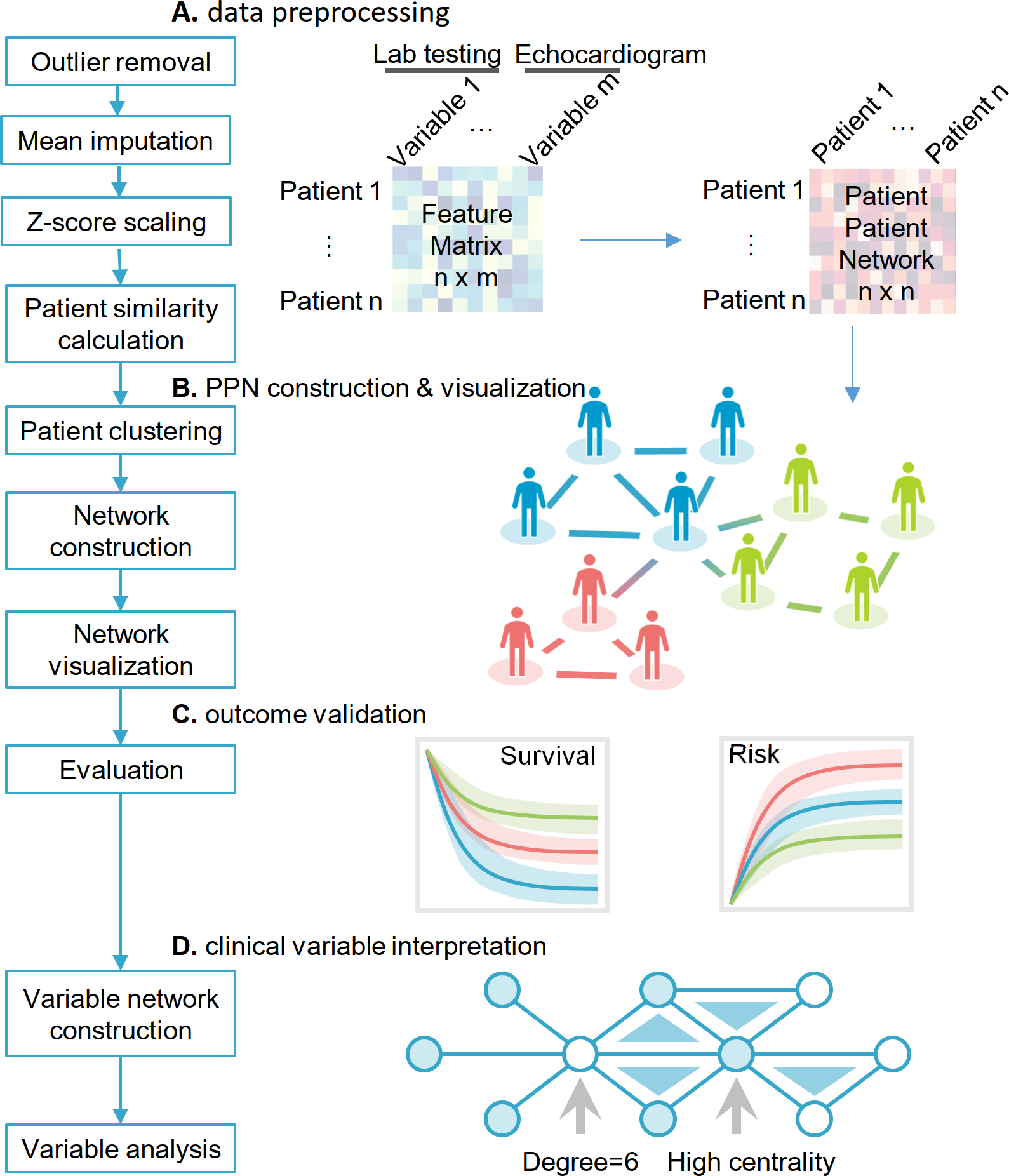
Cardiac risk stratification in cancer patients: A longitudinal patient–patient network analysis
Cardiovascular disease is a leading cause of death in general population and the second leading cause of mortality and morbidity in cancer survivors after recurrent malignancy in the United States. The growing awareness of cancer therapy–related cardiac dysfunction (CTRCD) has led to an emerging field of cardio-oncology; yet, there is limited knowledge on how to predict which patients will experience adverse cardiac outcomes. We aimed to perform unbiased cardiac risk stratification for cancer patients using our large-scale, institutional electronic medical records.
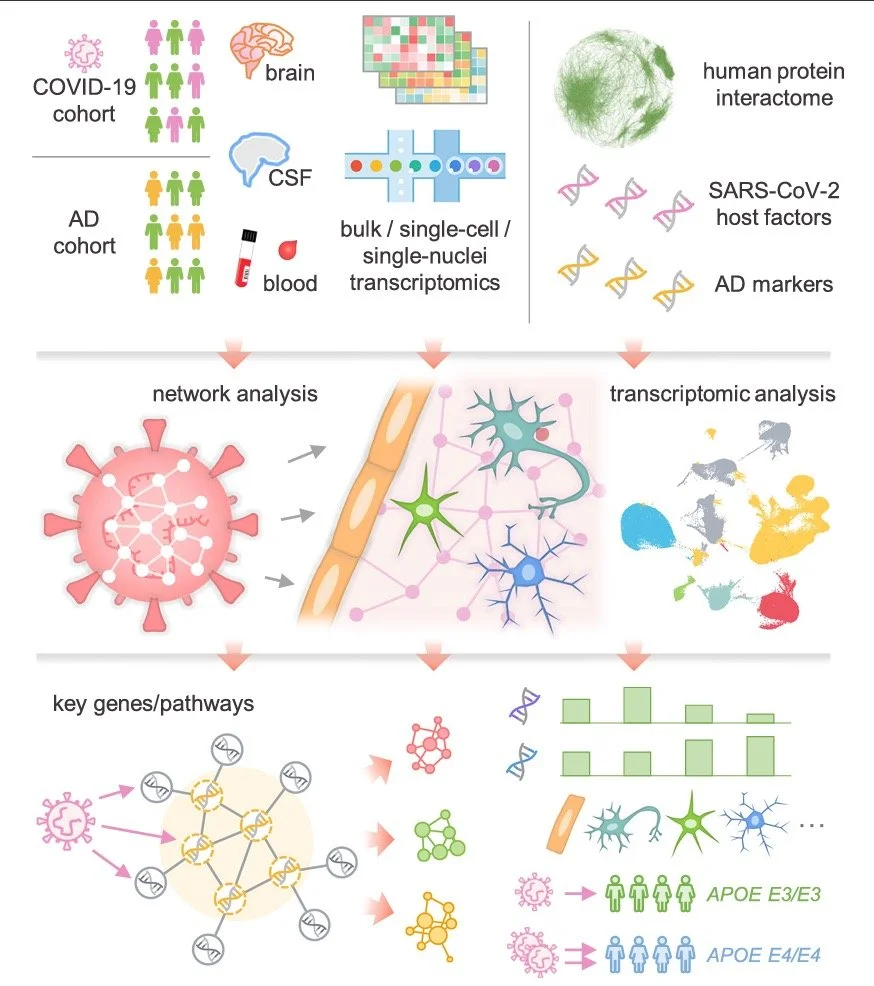
Network medicine links SARS-CoV-2/COVID19 infection to brain microvascular injury and neuroinflammation in dementia-like cognitive impairment
Dementia-like cognitive impairment is an increasingly reported complication of SARS-CoV-2 infection. However, the underlying mechanisms responsible for this complication remain unclear. A better understanding of causative processes by which COVID-19 may lead to cognitive impairment is essential for developing preventive and therapeutic interventions.
Network medicine framework for identifying drug-repurposing opportunities for COVID-19
The COVID-19 pandemic has highlighted the need to quickly and reliably prioritize clinically approved compounds for their potential effectiveness for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections. Here, we deployed algorithms relying on artificial intelligence, network diffusion, and network proximity, tasking each of them to rank 6,340 drugs for their expected efficacy against SARS-CoV-2. To test the predictions, we used as ground truth 918 drugs experimentally screened in VeroE6 cells, as well as the list of drugs in clinical trials that capture the medical community’s assessment of drugs with potential COVID-19 efficacy. We find that no single predictive algorithm offers consistently reliable outcomes across all datasets and metrics. This outcome prompted us to develop a multimodal technology that fuses the predictions of all algorithms, finding that a consensus among the different predictive methods consistently exceeds the performance of the best individual pipelines. We screened in human cells the top-ranked drugs, obtaining a 62% success rate, in contrast to the 0.8% hit rate of nonguided screenings. Of the six drugs that reduced viral infection, four could be directly repurposed to treat COVID-19, proposing novel treatments for COVID-19. We also found that 76 of the 77 drugs that successfully reduced viral infection do not bind the proteins targeted by SARS-CoV-2, indicating that these network drugs rely on network-based mechanisms that cannot be identified using docking-based strategies. These advances offer a methodological pathway to identify repurposable drugs for future pathogens and neglected diseases underserved by the costs and extended timeline of de novo drug development.
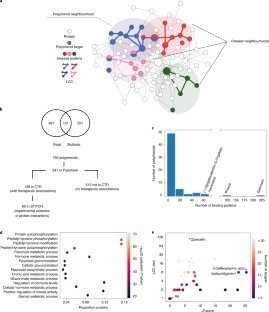
Network medicine framework shows that proximity of polyphenol targets and disease proteins predicts therapeutic effects of polyphenols
Polyphenols, natural products present in plant-based foods, play a protective role against several complex diseases through their antioxidant activity and by diverse molecular mechanisms. Here we develop a network medicine framework to uncover mechanisms for the effects of polyphenols on health by considering the molecular interactions between polyphenol protein targets and proteins associated with diseases. We find that the protein targets of polyphenols cluster in specific neighbourhoods of the human interactome, whose network proximity to disease proteins is predictive of the molecule’s known therapeutic effects. The methodology recovers known associations, such as the effect of epigallocatechin-3-O-gallate on type 2 diabetes, and predicts that rosmarinic acid has a direct impact on platelet function, representing a novel mechanism through which it could affect cardiovascular health. We experimentally confirm that rosmarinic acid inhibits platelet aggregation and α-granule secretion through inhibition of protein tyrosine phosphorylation, offering direct support for the predicted molecular mechanism. Our framework represents a starting point for mechanistic interpretation of the health effects underlying food-related compounds, allowing us to integrate into a predictive framework knowledge on food metabolism, bioavailability and drug interaction.
Comprehensive characterization of protein–protein interactions perturbed by disease mutations
Technological and computational advances in genomics and interactomics have made it possible to identify how disease mutations perturb protein–protein interaction (PPI) networks within human cells. Here, we show that disease-associated germline variants are significantly enriched in sequences encoding PPI interfaces compared to variants identified in healthy participants from the projects 1000 Genomes and ExAC. Somatic missense mutations are also significantly enriched in PPI interfaces compared to noninterfaces in 10,861 tumor exomes. We computationally identified 470 putative oncoPPIs in a pan-cancer analysis and demonstrate that oncoPPIs are highly correlated with patient survival and drug resistance/sensitivity. We experimentally validate the network effects of 13 oncoPPIs using a systematic binary interaction assay, and also demonstrate the functional consequences of two of these on tumor cell growth. In summary, this human interactome network framework provides a powerful tool for prioritization of alleles with PPI-perturbing mutations to inform pathobiological mechanism- and genotype-based therapeutic discovery.